Gencode exploration
Jun 4 2016 genomeGencode v19
I downloaded Gencode v19 at ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_human/release_19/gencode.v19.annotation.gtf.gz.
Genes
Number
Focusing on autosomes/X/Y, there are 57,783 “genes” of different types:
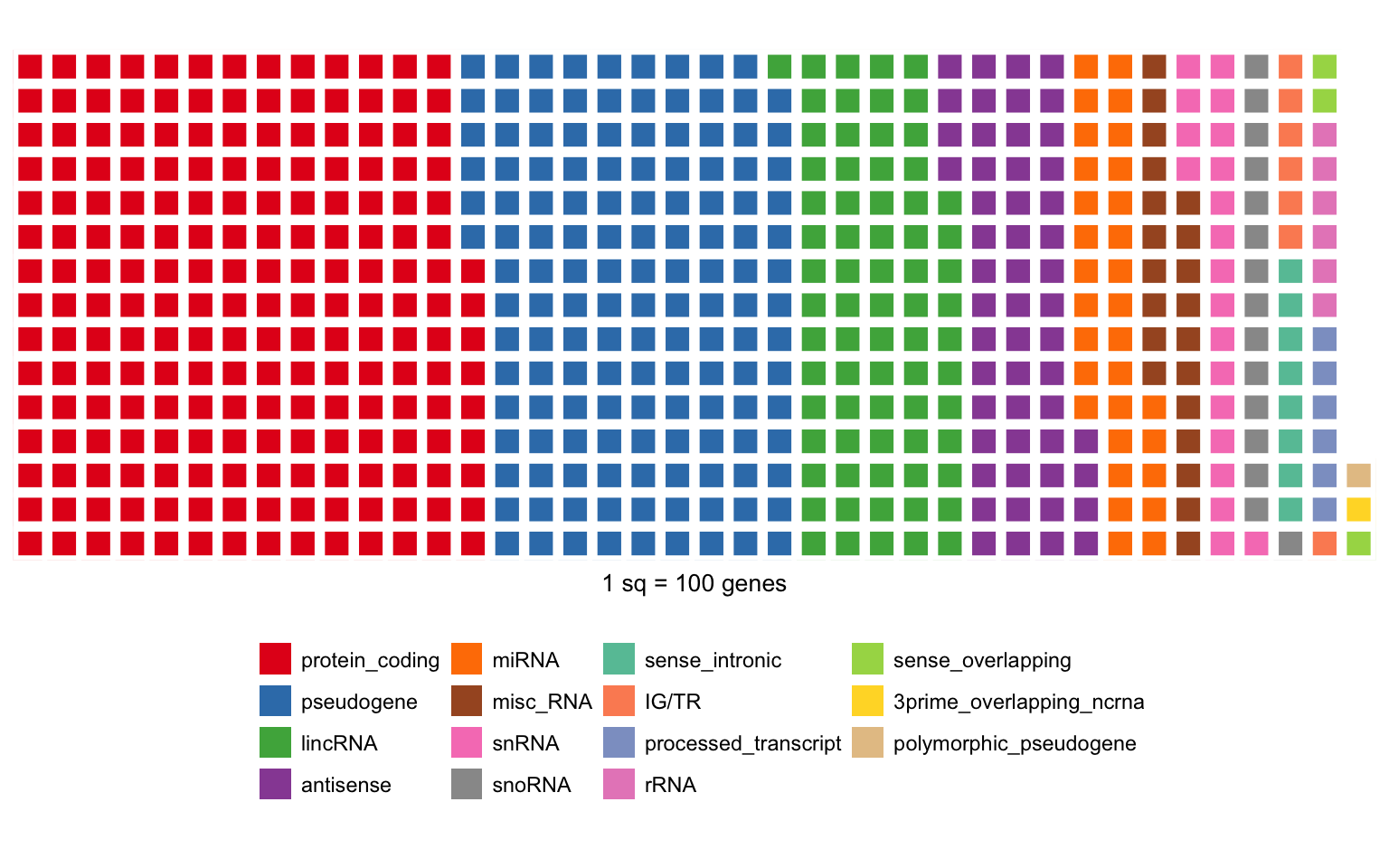
I merge the rare types into a other class and some RNAs.
| gene_type.f | n |
|---|---|
| protein_coding | 20332 |
| pseudogene | 13931 |
| other | 7417 |
| lincRNA | 7114 |
| RNA | 5934 |
| miRNA | 3055 |
Size
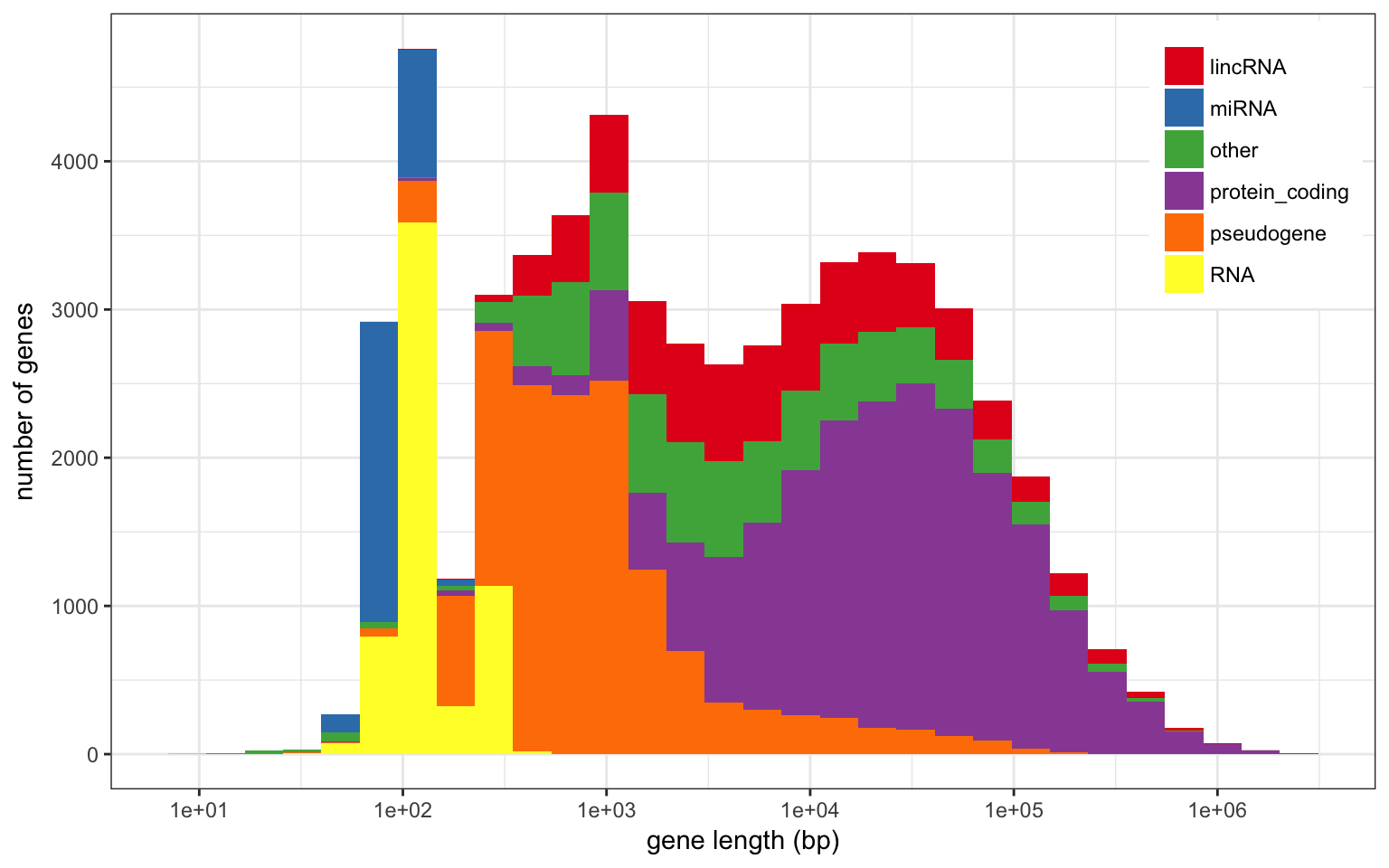
The largest annotated genes span more than 2 Mbp:
| seqnames | gene_name | gene_type | size.Mbp |
|---|---|---|---|
| chr7 | CNTNAP2 | protein_coding | 2.304638 |
| chr9 | PTPRD | protein_coding | 2.298478 |
| chrX | DMD | protein_coding | 2.241765 |
| chr3 | LSAMP | protein_coding | 2.194861 |
| chr11 | DLG2 | protein_coding | 2.172912 |
The smallest protein-coding annotated genes less than 100 bp:
| seqnames | gene_name | gene_type | size.bp |
|---|---|---|---|
| chr2 | AC011308.1 | protein_coding | 59 |
| chr12 | AC055736.1 | protein_coding | 66 |
| chr16 | PIH1 | protein_coding | 93 |
| chr2 | AC012360.2 | protein_coding | 96 |
| chr5 | AC008914.1 | protein_coding | 102 |
Density
Using non-overlapping windows of 1 Mb the gene density looks like this:
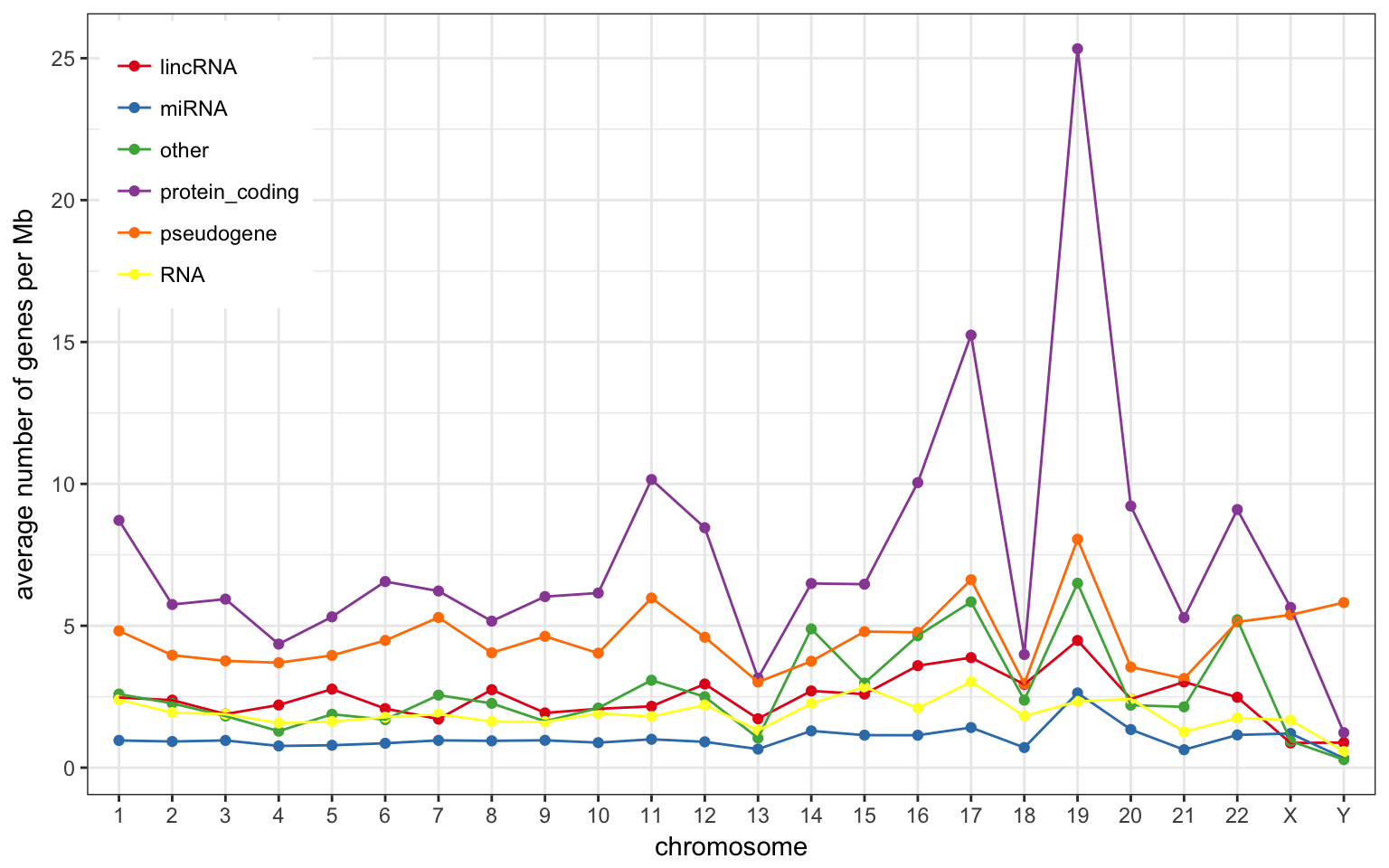
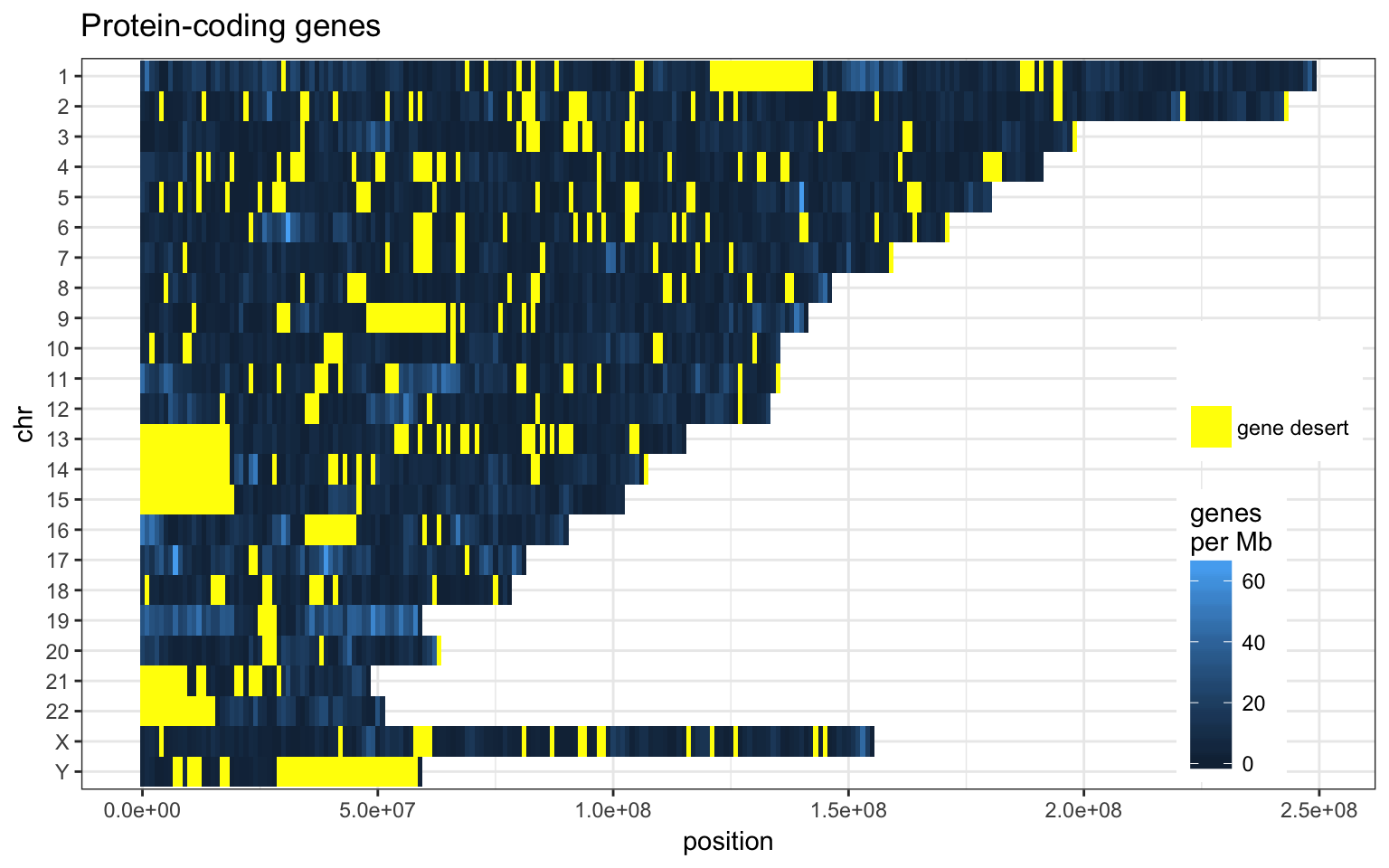
- Chr 19, 17 and 11 have more protein-coding genes than the rest.
- Chr Y has more pseudogene compared to other classes.
Exons
Number
Focusing on autosomes/X/Y, there are 1,196,256 “exons” from different types of genes:
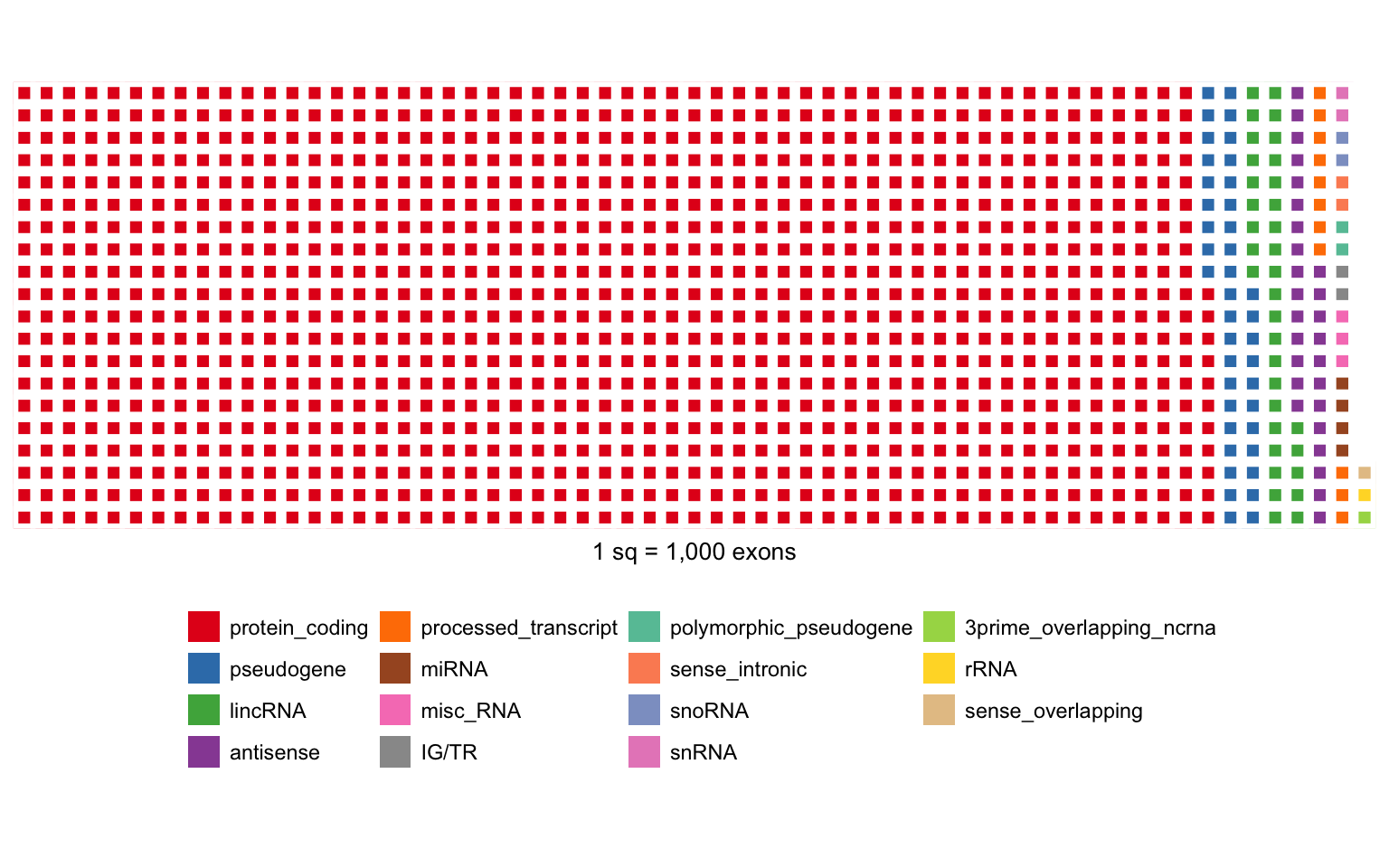
For the rest of the analysis, I use only exons from protein-coding genes.
Number per gene
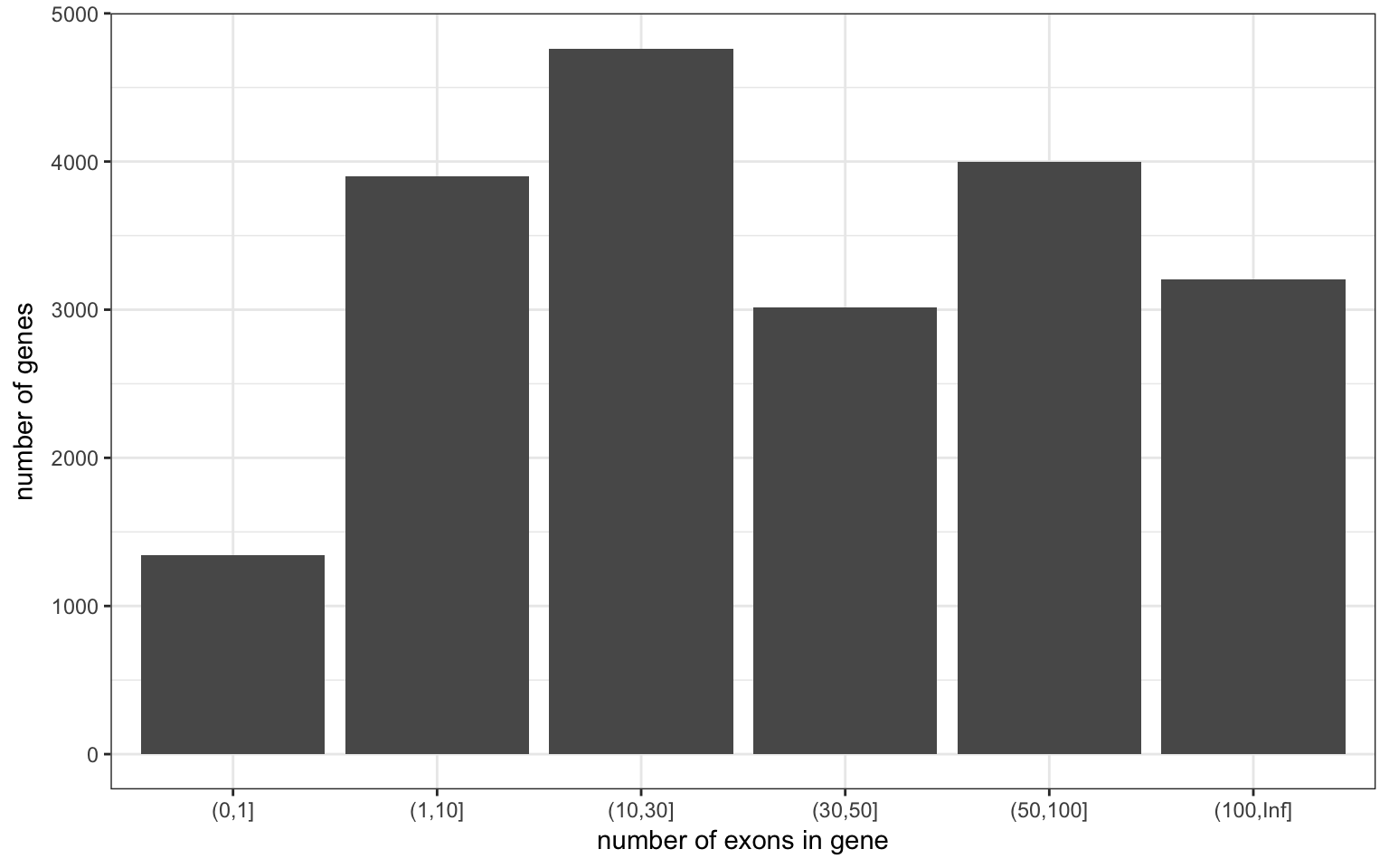
| mean.nb.exon | median.nb.exon | max.nb.exon |
|---|---|---|
| 52.9 | 31 | 1696 |
The gene with the most exons is the Titin gene:
| gene_name | exon |
|---|---|
| TTN | 1696 |
| SYNE1 | 1377 |
| NEB | 1225 |
| CACNA1G | 1139 |
| CACNA1C | 1098 |
Size
The average exon size is 232 bp.
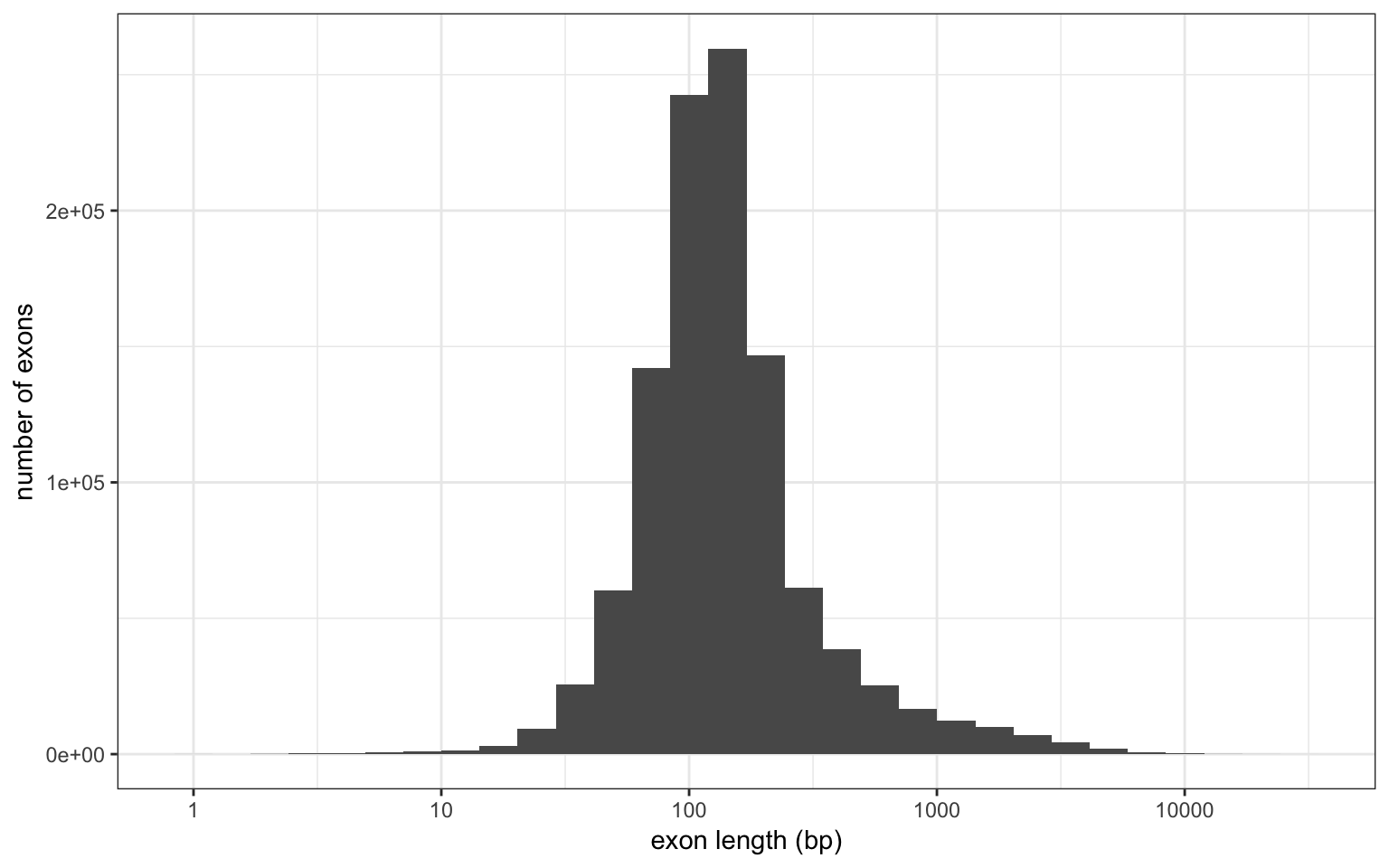
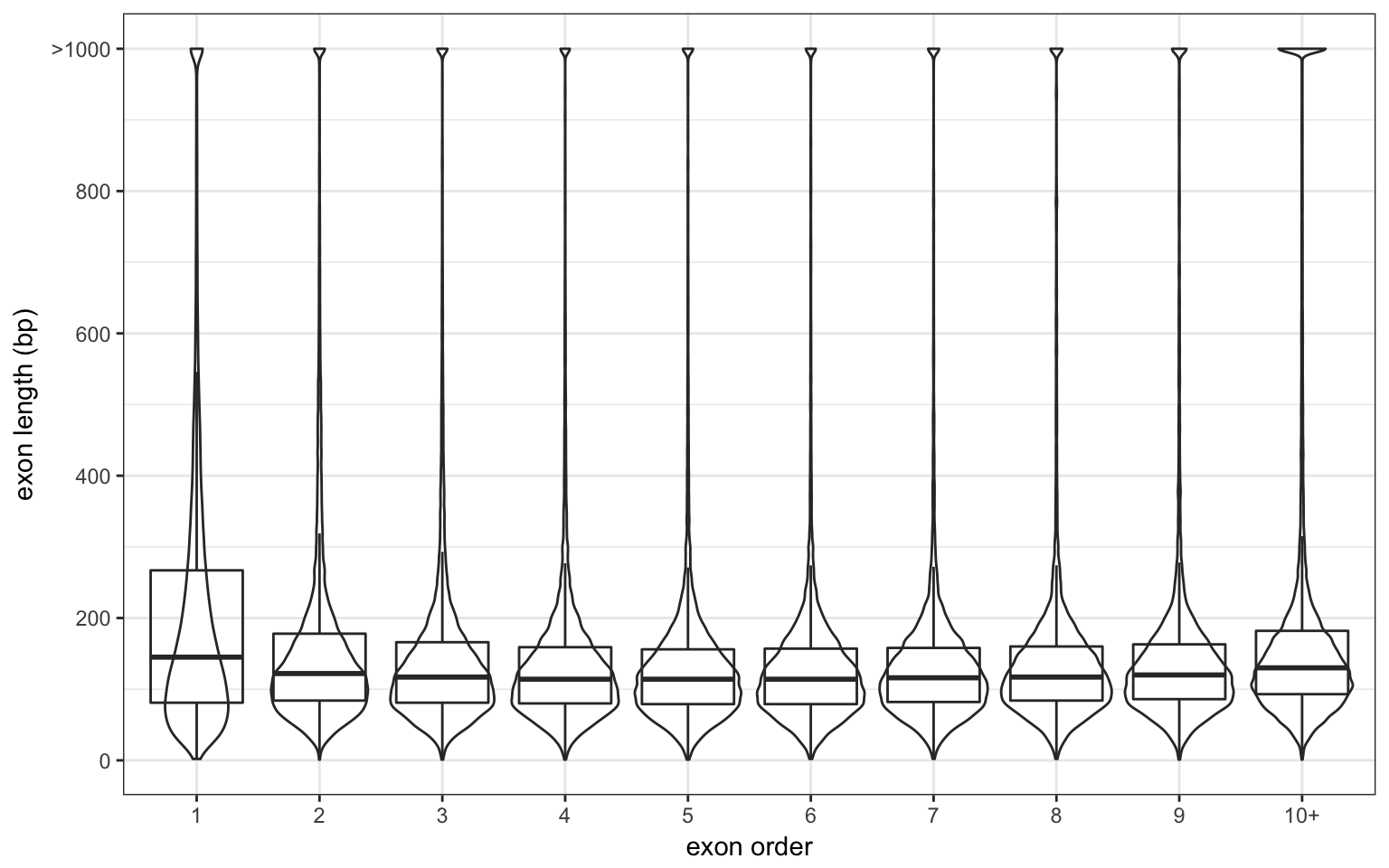
The first exon seems to be slightly larger than the others. I used genes with at least 10 exons to be sure it’s not due to large single-exon genes.
The largest annotated exons are more than 20 Kbp long:
| seqnames | gene_name | size.kb |
|---|---|---|
| chr8 | TRAPPC9 | 29.06 |
| chr5 | MCC | 27.20 |
| chr12 | GRIN2B | 24.41 |
| chr19 | MUC16 | 21.69 |
| chr2 | ABI2 | 20.55 |
The smallest are just 1 bp !?
| seqnames | gene_name | size.bp |
|---|---|---|
| chr2 | ALK | 1 |
| chr3 | ACAD11 | 1 |
| chr4 | PPA2 | 1 |
| chr5 | PAM | 1 |
| chr5 | GALNT10 | 1 |