Segmental duplication exploration
Oct 20 2016 genomeSegmental Duplications (SD)
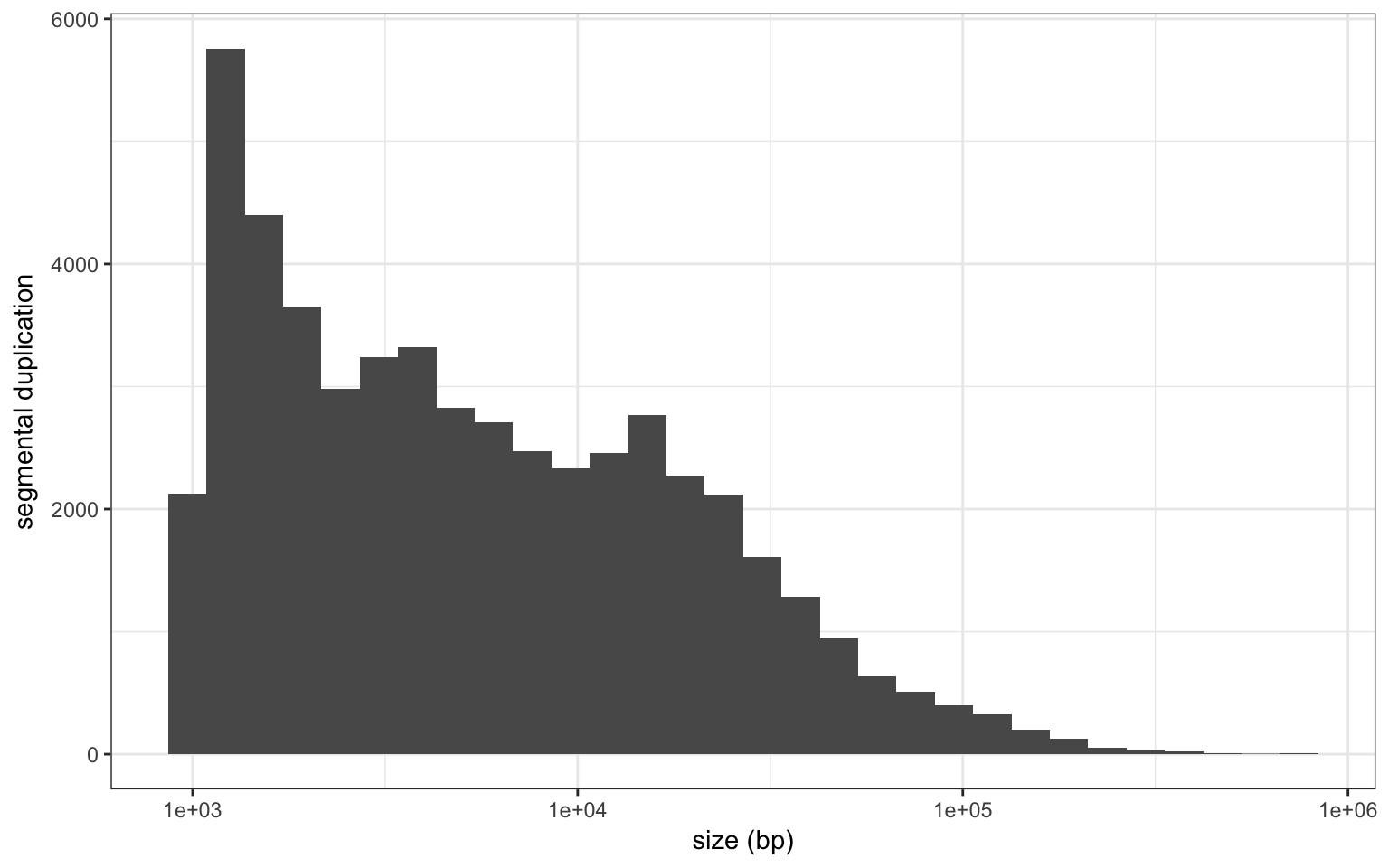
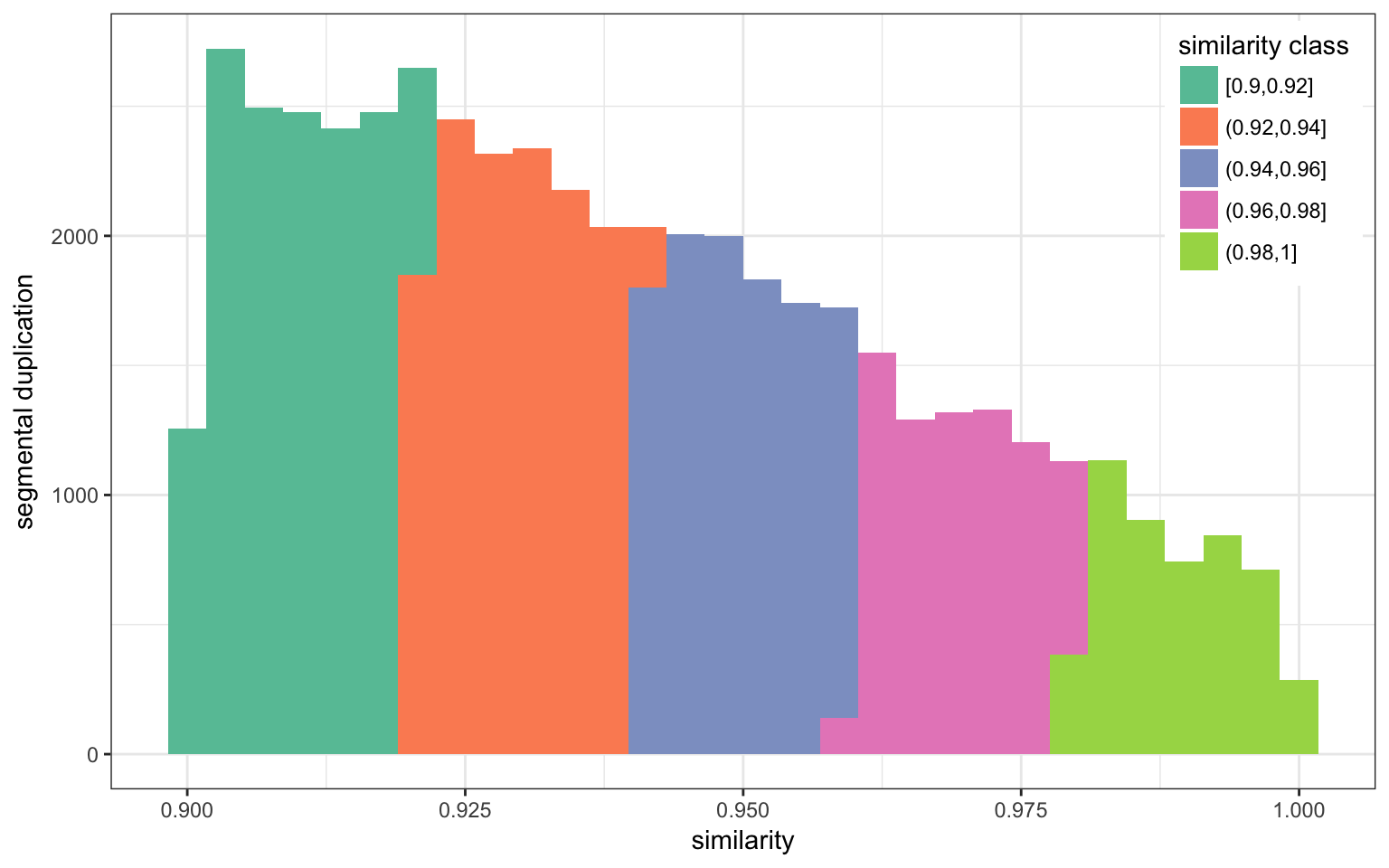
I downloaded the segmental duplication annotation for hg19 from UCSC. There are 51599 annotated SD. They are defined as regions larger than 1 Kbp with at least 90% similarity with another region in the genome.
Segmental duplication regions
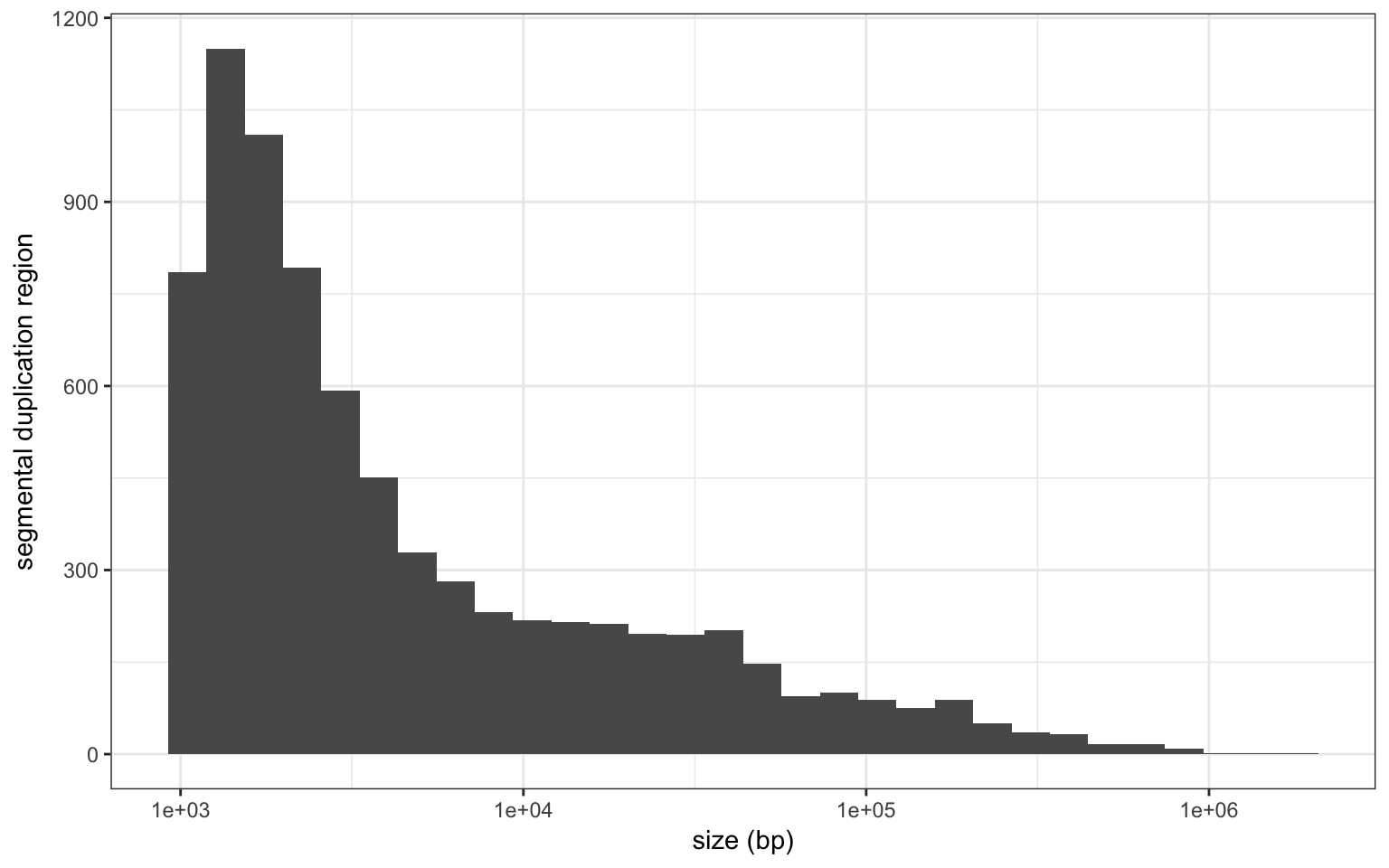
Many SD are nested of located next to each other. I merge overlapping SDs (or located at <10 bp) to create SD regions, i.e. longer stretch of the genome overlapping SDs.
There are 7620 SD regions, that account for 166.1 Mbp of the genome.
Size distribution
Similarity
Chromosome distribution
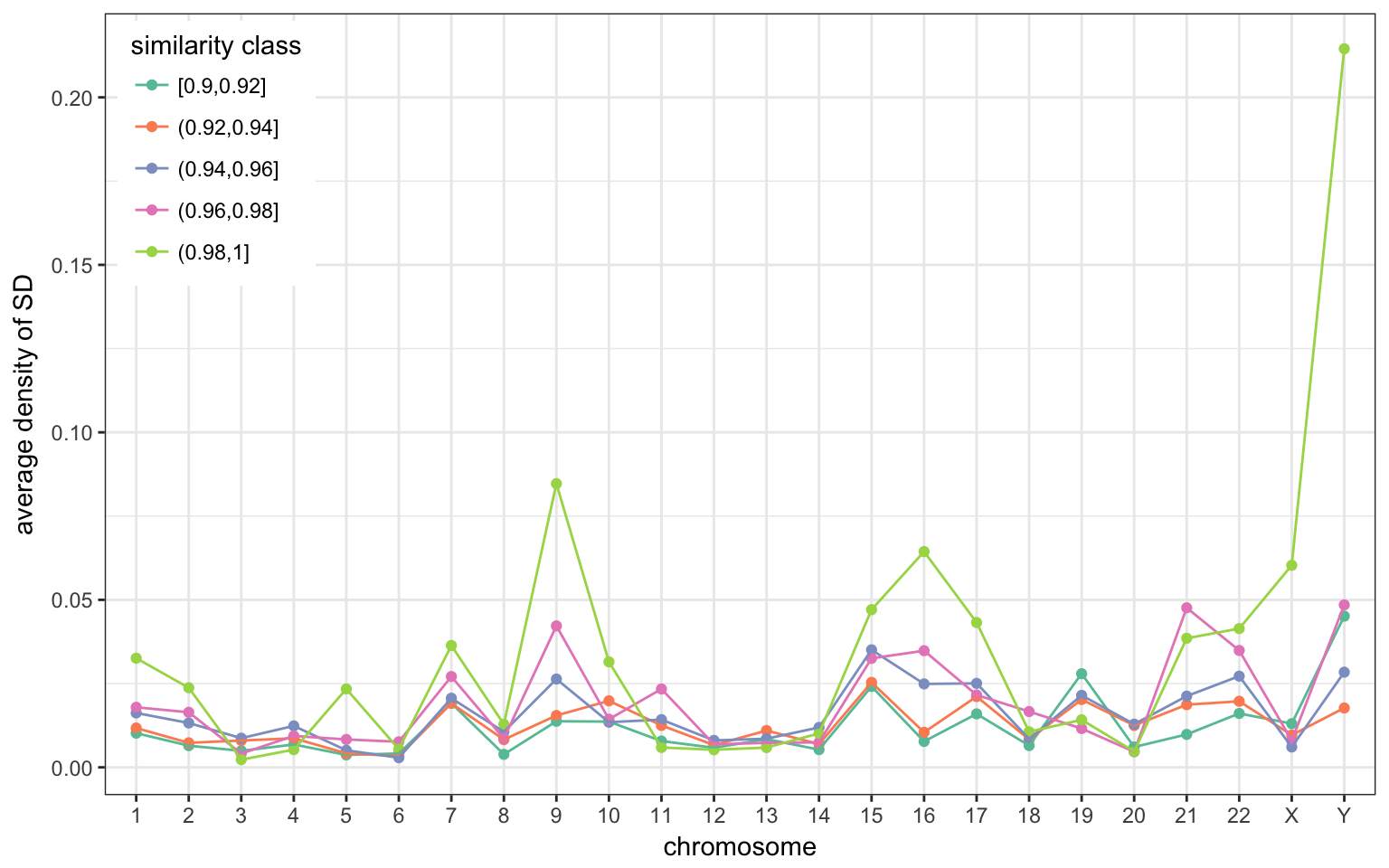
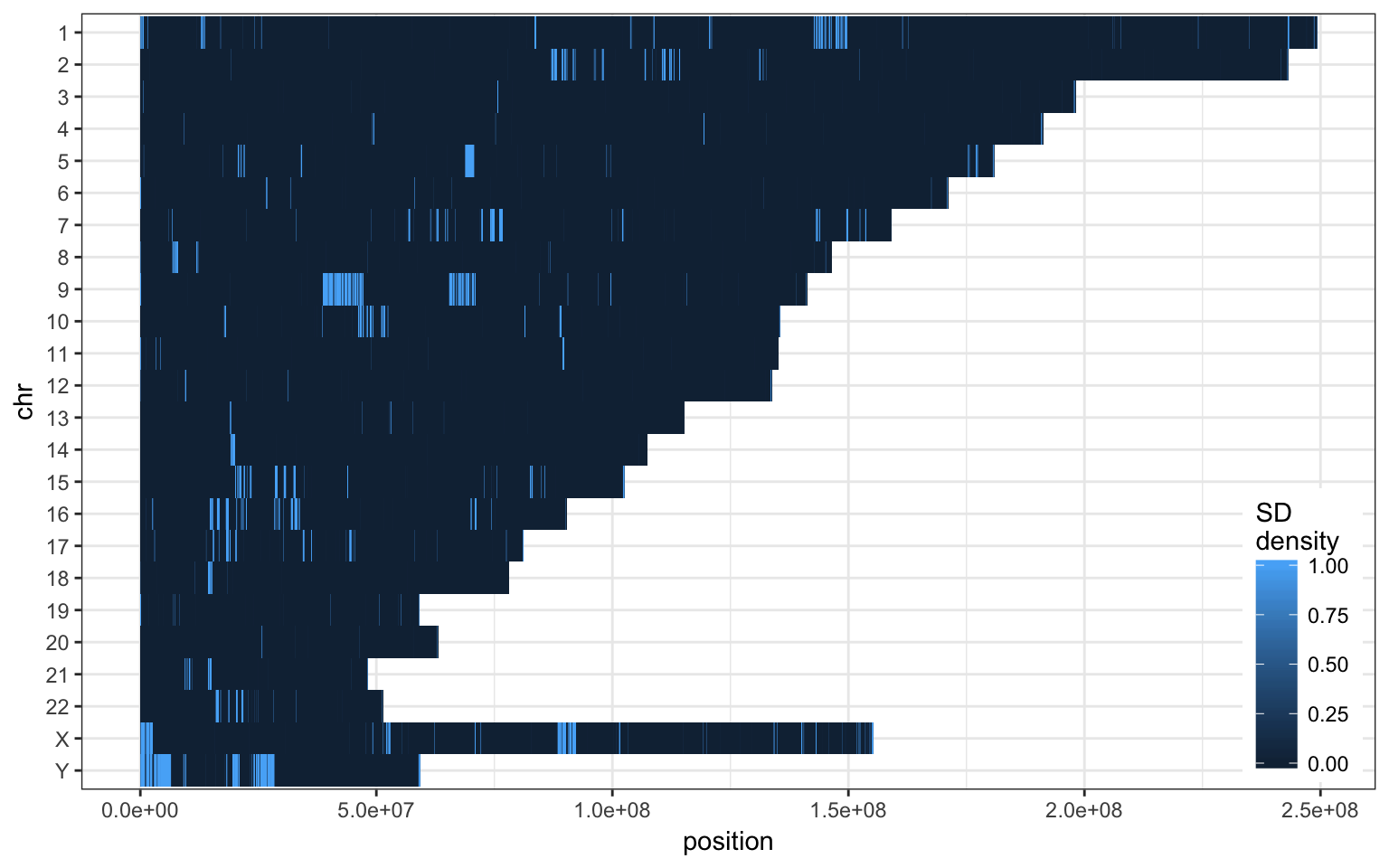
A few chromosomes are more enriched in SD. Some have long stretches of SD, e.g. chr 9 or chr Y. These peaks are mostly created with very recent/similar SDs.
Distance to the other segment
| otherSeg | segdup | segdup.prop |
|---|---|---|
| different chr | 32740 | 0.635 |
| same chr | 18859 | 0.365 |
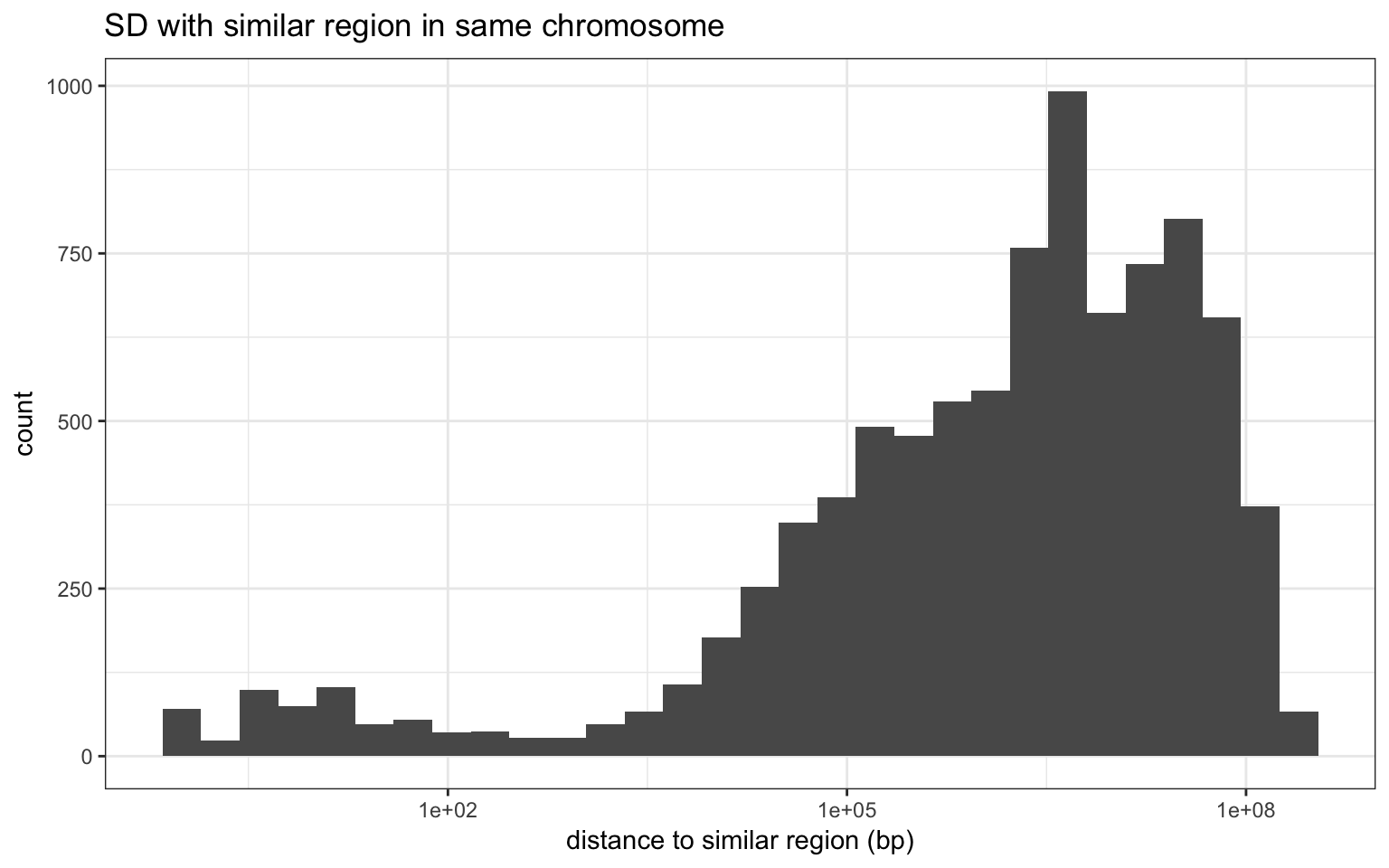
For the majority of SDs, the similar region is in a different chromosome. For the others the majority are far from each other.
Gene content
I downloaded Gencode v19 at ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_human/release_19/gencode.v19.annotation.gtf.gz.
Around a thousand protein-coding genes are completely within SD regions.
| gene_type | gene |
|---|---|
| pseudogene | 3968 |
| protein_coding | 958 |
| lincRNA | 641 |
| miRNA | 301 |
| antisense | 181 |
| snRNA | 179 |
| IG_V_pseudogene | 97 |
| misc_RNA | 95 |
| IG_V_gene | 86 |
| rRNA | 62 |
| processed_transcript | 60 |
| sense_intronic | 29 |
| TR_V_gene | 25 |
| snoRNA | 23 |
| IG_D_gene | 15 |
| IG_C_gene | 9 |
| polymorphic_pseudogene | 8 |
| TR_V_pseudogene | 7 |
| TR_J_gene | 5 |
| sense_overlapping | 4 |
| IG_C_pseudogene | 2 |
| IG_J_gene | 2 |
| TR_C_gene | 2 |
Gene families
A naive way of looking for gene families is to cluster the gene names. I also remove any trailing numbers in the gene name.
| example | n |
|---|---|
| PRAMEF | 21 |
| USP17L | 21 |
| ZNF | 15 |
| GOLGA8I | 13 |
| NBPF | 12 |
| CT47B | 12 |
| POTEF | 11 |
| OR2T | 10 |
Gene Ontology
| Description | GeneRatio | qvalue | |
|---|---|---|---|
| GO:0050907 | detection of chemical stimulus involved in sensory perception | 57/509 | 0.0000000 |
| GO:0009593 | detection of chemical stimulus | 58/509 | 0.0000000 |
| GO:0007606 | sensory perception of chemical stimulus | 59/509 | 0.0000000 |
| GO:0007608 | sensory perception of smell | 53/509 | 0.0000000 |
| GO:0050911 | detection of chemical stimulus involved in sensory perception of smell | 51/509 | 0.0000000 |
| GO:0050906 | detection of stimulus involved in sensory perception | 57/509 | 0.0000000 |
| GO:0042742 | defense response to bacterium | 23/509 | 0.0002887 |
| GO:0098542 | defense response to other organism | 35/509 | 0.0002887 |
| GO:0071346 | cellular response to interferon-gamma | 17/509 | 0.0002992 |
| GO:0006805 | xenobiotic metabolic process | 14/509 | 0.0002992 |
| GO:0071466 | cellular response to xenobiotic stimulus | 14/509 | 0.0004487 |
| GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein | 7/509 | 0.0004487 |
| GO:0002323 | natural killer cell activation involved in immune response | 8/509 | 0.0005222 |
| GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein | 7/509 | 0.0005486 |
| GO:0009410 | response to xenobiotic stimulus | 14/509 | 0.0007752 |
| GO:0034341 | response to interferon-gamma | 17/509 | 0.0012694 |
| GO:0006749 | glutathione metabolic process | 10/509 | 0.0014056 |
| GO:0042501 | serine phosphorylation of STAT protein | 7/509 | 0.0019334 |
| GO:0016579 | protein deubiquitination | 14/509 | 0.0035460 |
| GO:0060337 | type I interferon signaling pathway | 11/509 | 0.0040381 |
| GO:0071357 | cellular response to type I interferon | 11/509 | 0.0040381 |
| GO:0006342 | chromatin silencing | 13/509 | 0.0046589 |
| GO:0034340 | response to type I interferon | 11/509 | 0.0058697 |
| GO:0030101 | natural killer cell activation | 10/509 | 0.0090640 |
| GO:0070646 | protein modification by small protein removal | 14/509 | 0.0114298 |
| GO:0009812 | flavonoid metabolic process | 6/509 | 0.0138310 |
| GO:0033559 | unsaturated fatty acid metabolic process | 12/509 | 0.0138310 |
| GO:0000042 | protein targeting to Golgi | 5/509 | 0.0138310 |
| GO:0019373 | epoxygenase P450 pathway | 5/509 | 0.0138310 |
| GO:0045814 | negative regulation of gene expression, epigenetic | 13/509 | 0.0138310 |
| GO:0060338 | regulation of type I interferon-mediated signaling pathway | 7/509 | 0.0139433 |
| GO:0006690 | icosanoid metabolic process | 11/509 | 0.0144589 |
| GO:0009813 | flavonoid biosynthetic process | 5/509 | 0.0165692 |
| GO:0006959 | humoral immune response | 15/509 | 0.0202359 |
| GO:0002228 | natural killer cell mediated immunity | 8/509 | 0.0226821 |
| GO:0006334 | nucleosome assembly | 13/509 | 0.0227629 |
| GO:0006968 | cellular defense response | 8/509 | 0.0232698 |
| GO:0052696 | flavonoid glucuronidation | 5/509 | 0.0232698 |
| GO:0072600 | establishment of protein localization to Golgi | 5/509 | 0.0232698 |
| GO:0043330 | response to exogenous dsRNA | 7/509 | 0.0234523 |
| GO:0001906 | cell killing | 11/509 | 0.0249099 |
| GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste | 6/509 | 0.0322850 |
| GO:0052695 | cellular glucuronidation | 5/509 | 0.0331023 |
| GO:0042737 | drug catabolic process | 4/509 | 0.0378567 |
| GO:0000301 | retrograde transport, vesicle recycling within Golgi | 5/509 | 0.0389140 |
| GO:0031497 | chromatin assembly | 13/509 | 0.0483266 |
| Description | GeneRatio | qvalue | |
|---|---|---|---|
| GO:0004984 | olfactory receptor activity | 51/488 | 0.0000000 |
| GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 8/488 | 0.0000499 |
| GO:0005132 | type I interferon receptor binding | 7/488 | 0.0000499 |
| GO:0008391 | arachidonic acid monooxygenase activity | 6/488 | 0.0002238 |
| GO:0008392 | arachidonic acid epoxygenase activity | 6/488 | 0.0002238 |
| GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity | 14/488 | 0.0002238 |
| GO:0101005 | ubiquitinyl hydrolase activity | 14/488 | 0.0002238 |
| GO:0008395 | steroid hydroxylase activity | 8/488 | 0.0002715 |
| GO:0019825 | oxygen binding | 9/488 | 0.0004195 |
| GO:0019783 | ubiquitin-like protein-specific protease activity | 14/488 | 0.0005119 |
| GO:0003823 | antigen binding | 11/488 | 0.0007524 |
| GO:0004497 | monooxygenase activity | 12/488 | 0.0008659 |
| GO:0070330 | aromatase activity | 6/488 | 0.0009112 |
| GO:0020037 | heme binding | 13/488 | 0.0029793 |
| GO:0048020 | CCR chemokine receptor binding | 7/488 | 0.0030635 |
| GO:0019864 | IgG binding | 4/488 | 0.0038146 |
| GO:0046906 | tetrapyrrole binding | 13/488 | 0.0050261 |
| GO:0042605 | peptide antigen binding | 5/488 | 0.0078319 |
| GO:0005506 | iron ion binding | 13/488 | 0.0176137 |
| GO:0004364 | glutathione transferase activity | 5/488 | 0.0236924 |
| GO:0005125 | cytokine activity | 16/488 | 0.0236924 |
| GO:0008234 | cysteine-type peptidase activity | 14/488 | 0.0236924 |
| GO:0042379 | chemokine receptor binding | 7/488 | 0.0364788 |
| GO:0015020 | glucuronosyltransferase activity | 5/488 | 0.0364788 |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 12/488 | 0.0364788 |
| GO:0019865 | immunoglobulin binding | 4/488 | 0.0422110 |
| GO:0008009 | chemokine activity | 6/488 | 0.0435900 |
| Description | GeneRatio | qvalue | |
|---|---|---|---|
| GO:0045095 | keratin filament | 16/589 | 0.0000242 |
| GO:0005882 | intermediate filament | 19/589 | 0.0042923 |
| GO:0042611 | MHC protein complex | 6/589 | 0.0057256 |
| GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane | 6/589 | 0.0074647 |
| GO:0098553 | lumenal side of endoplasmic reticulum membrane | 6/589 | 0.0074647 |
| GO:0045111 | intermediate filament cytoskeleton | 19/589 | 0.0171090 |
| GO:0000786 | nucleosome | 11/589 | 0.0178803 |
| GO:0044815 | DNA packaging complex | 11/589 | 0.0258667 |
| GO:0012507 | ER to Golgi transport vesicle membrane | 7/589 | 0.0434286 |
| GO:0072562 | blood microparticle | 11/589 | 0.0438412 |